Tools
1. FillMD@Mks: a tool to replace genotyping errors with missing data
Genotyping errors coming from the file "events_summary.log" can be filled with missing data ("-") after that the consistency of marker genotyping information has been checked along generations of selection.
Select Tools > FillMD@Mks... from the menu bar (see below).
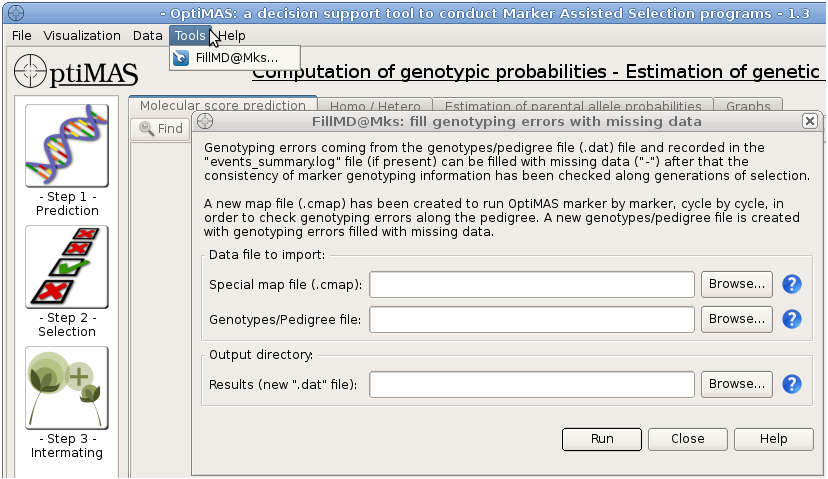
Figure 32: data set importation to run the FillMd@Mks tool
Loading input/output files using the browser:
Map file: path to a new genetic map/QTL position input file (.cmap). This file has been created at the end of a previous run of OptiMAS in order to re-run OptiMAS marker by marker and localize genotyping errors at individual marker position.
Genotype file: path to the genotype/pedigree file (.dat). This is the same file used to run OptiMAS the first time.
Output directory: path to the folder where a new genotype/pedigree file (.dat) with inconsistent genotyping data replaced by missing data ("-") will be stored. Note that your output directory should not be in the "Program Files" folder or other specific directories with administrator privileges.
Click on the “Run” button to create this new genotype/pedigree file (new_file.dat). Use this new .dat file to re-run OptiMAS.