
Mining genetic diversity for tomorrow’s agriculture
Genetic diversity is the key ingredient
fueling gains during plant breeding programs. Thus,
understanding the structure of a germplasm’s
genetic diversity as well as the factors shaping it pave
the way for crop improvement. During my thesis, I
utilized quantitative modeling and bioinformatic
approaches to study both meiotic recombination, a
factor driving genome reshuffling, and the genetic
diversity of two important crops, tomato and peanut.
For meiotic recombination, individual
genomic/epigenomic features have weak predictive
power regarding the distribution of crossovers in
Arabidopsis thaliana. Instead, a summarized
epigenetic status, referring to 10 chromatin states, is
able to reveal the associated landscape rather well.
Furthermore, I found that intermediate levels of
single nucleotide polymorphisms (SNPs) between
homologs leads to more crossovers compared to the
case of near identical sequences, and that intergenic
regions of size less than 1.5 kb tend to suppress
crossovers. Taken together, I integrated these effects
into a quantitative model that can predict
recombination landscapes and that reproduces much
of the variation seen in the experimental crossover
data.
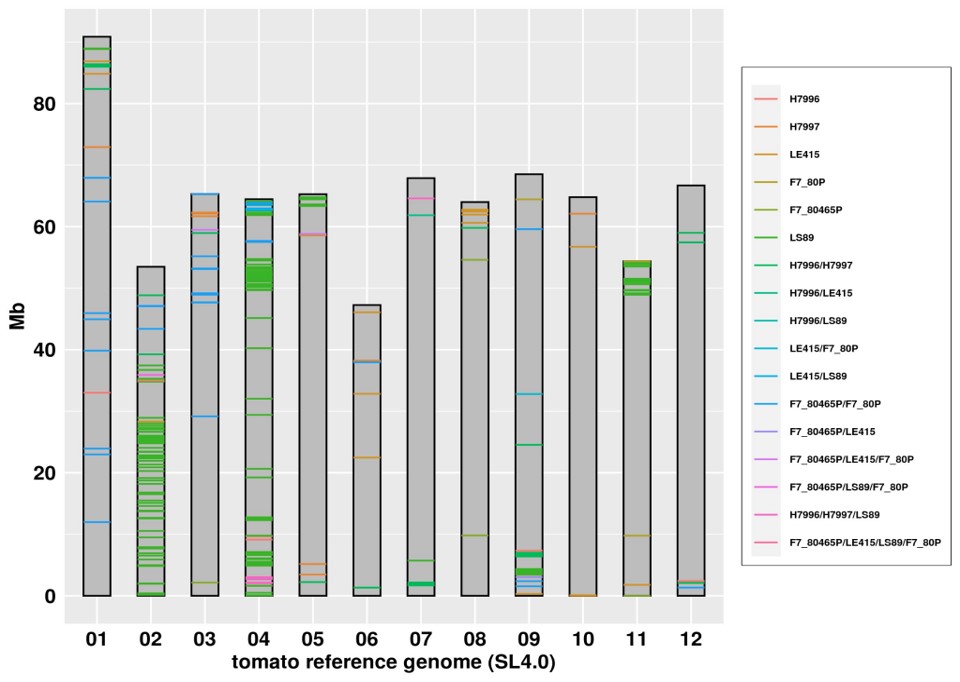
Moving on to the cultivated tomato, one of its most
destructive diseases is Bacterial wilt (BW). I used the
whole genome sequence data of six BW resistant
and nine BW susceptible lines to identify
polymorphisms specific to resistant lines. Among
resistant-specific polymorphisms affecting 385
genes, the marker Bwr3.2dCAPS located in the Asc
(Solyc03g114600.4.1) was shown to be significantly
associated with the BW resistance but nevertheless
it does not fully explain the resistance phenotype.
Lastly, I assessed the genetic diversity of cultivated
peanuts in Taiwan by the restriction site associated
DNA (RAD) approach using 31 accessions. My
results indicate that worldwide accessions have
greater genetic diversity than local accessions,
suggesting that novel genetic resources should be
introduced into the present breeding programs for
enhancing the genetic diversity. These successive
investigations, motivated respectively by
fundamental biology and by applied breeding
science, provide new insights that can help future
strategies for crop improvement.
peanut genetic diversity tomato resistance gene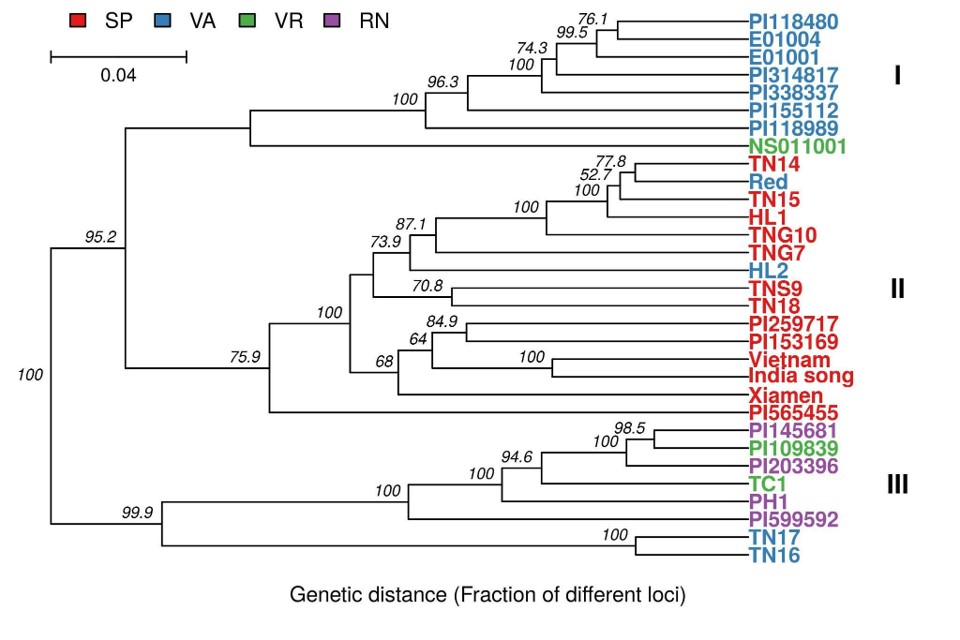
Jury
Beth Rowan, Cécile Fairhead, Martin Howard, Mathias Lorieux and Piotr Ziółkowski.