|
Research interests
My
interests include evolution of the plant genome with a particular
focus on interspecific hybridization and polyploidy, and dynamics
of transposable elements (TEs) as well as their epigenetic control by
small non coding RNAs.
TEs account
for most nuclear DNA in higher plants and might be a source of genetic
variability, because they replicate and reinsert at multiple sites in
the genome and may act as mutagenic agents. Moreover, TEs are activated
under stress conditions and thus contribute to genome adaptation.
Polyploidization which consists in the reuniting of two (or more)
genomes in a common nucleus after long periods of divergence can be
considered as a state of genome stress that could precipitate the
activation of TEs. Assuming that nearly all angiosperms have
experienced at least one episode of polyploidization during their
evolution, the question of how TEs can be involved in
the formation of a polyploid species is a major task.
Brassica is an agriculturally
important genus that provides a wide range of diploid and allotetraploid
species, and is particularly appropriate for studying the evolution of TEs
during speciation and polyploidization.
Part of my
research project is dedicated to the analysis of the impact of a recent
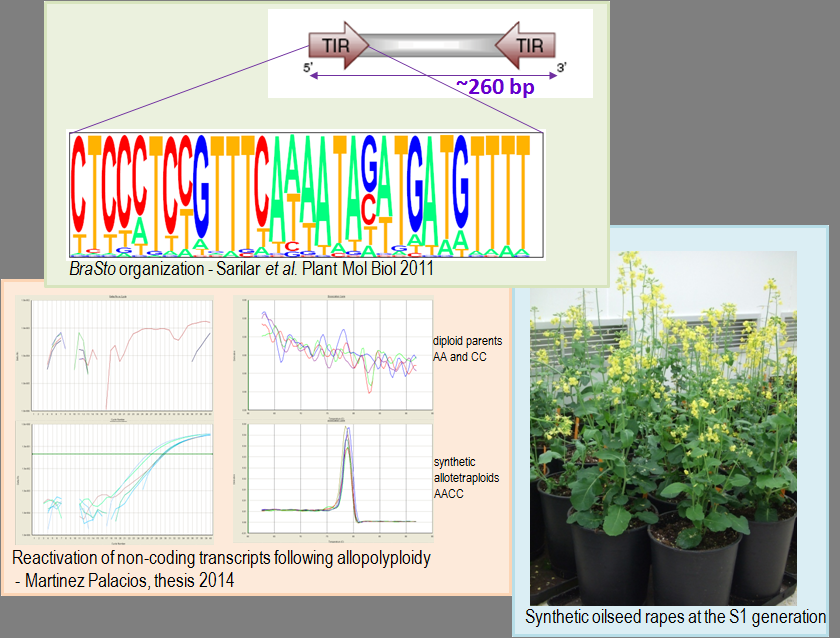
polyploidization event on the activation of TEs by studying the evolution of Brassica
-specific TE families (retrotransposons, MITEs, CACTA elements) in newly synthesized
Brassica napus lines. The activation of
TEs is surveyed at both the
structural and functional levels, using a combination of molecular
and bioinformatic approaches. This represented part of Véronique Sarilar's PhD project (2008-2011).
Another part of my project deals with the role of
small non coding RNAs in regulating the neo-allopolyploid Brassica
genome during its formation. We specifically focus our analysis on the
role of snRNAs in controlling non-coding components of the genome
(including TEs). Paulina
Martinez Palacios' PhD work was dedicated to the global analysis of the
mobilisation of snRNAs following allopolyploidization in the newly
synthesized B. napus lines (2010-2014).
Finally, I am interested in the proteomic
and molecular characterization of gene expression and its regulation
following a recent allopolyploidy event: comparative proteomics is
performed on early (S0-S1) and more advanded (S5) generations of the same newly synthesized
B. napus lines
to identify modifications of gene regulation occuring at the protein
level. Anne Marmagne was involved in this project during her
postdoc (2006-2008).
In relation with my interest in interspecific hybridization as a major process in plant evolution, I also collaborate with Yves Vigouroux, Dynadiv - IRD Montpellier, in supervising Roland Akakpo's PhD project (ED 'Sciences du Végétal : du gène à l'écosystème', Université Paris-Sud, Orsay). Our main objective is there to clarify the genetic relationships between three Dioscorea species, with D. abyssinica and D. praehensilis considered as the progenitors of the cultivated yam species D. rotundata. At GQE-Le Moulon, and in collaboration with Florian Maumus and Hadi Quesneville, URGI - INRA Versailles,
we analyse the diversity and organisation of TEs among the three
species, by exploiting NGS data; our objective is to identify TE
families at the origins of genome differentiation between the three
species.
Positions and Education
- 2013: HDR defense, Université Paris-Sud. « Allopolyploïdie et éléments transposables : contribution à l'étude de la dynamique évolutive du génome des Brassica » / « Allopolyploidy and transposable elements: contribution to the study of the evolutionary dynamics of the Brassica genome »
- Transposable elements
and evolution of the
Brassica
genome
- Allopolyploidization and activation of TEs: structural and
functional analysis of newly synthesized
B.
napus
allotetraploids (Véronique Sarilar, PhD student / coll. Philippe Brabant, Johann Joets - UMR GQE-Le Moulon)
-
2003-2004:
Postdoctoral position,
UMR Amélioration des Plantes et
Biotechnologies Végétales
, INRA-Agrocampus Rennes - M. Manzanares-Dauleux
Molecular and functional characterization
of genetic components of resistance to clubroot (
Plasmodiophora brassicae
) in
Arabidopsis thaliana
and
Brassica napus
Alix K.
, Lariagon C., Delourme R., Manzanares-Dauleux M.J. 2007. Exploiting natural genetic
diversity and mutant resources of
Arabidopsis thaliana
to study the
A. thaliana
-
Plasmodiophora brassicae
interaction.
Plant Breeding 126: 218-221
.
-
2002:
Temporary position,
UMR Amélioration des Plantes et
Biotechnologies Végétales
, INRA-Agrocampus Rennes - A.-M. Chèvre
-
Monitoring introgression
of
resistance
to
blackleg in
Brassica napus
from the B genome of mustards using
genomic
in situ
hybridisation (GISH)
-
Analysis of the genomic constitution of oilseed
rape - wild radish hybrids by GISH in a context of gene flow assessment
-
2001:
Postdoctoral fellow (OECD) in
Pat Heslop-Harrison's lab
, Department of Biology, Univ. Leicester, Leicester /United Kingdom
The diversity of retrotransposons in
Brassica
: understanding their phylogeny and evolution
- 1999-2000:
Postdoctoral position, UMR Diversité et Génome des
Plantes Cultivées, INRA-Montpellier - P. Roumet, J.
David
-
Implication of peroxidases in clearness of pasta:
development of S-SAP markers in durum wheat
-
Role of the
Ph1
gene in the intergenomic recombination rate in
Triticum durum
×
Aegilops ovata
amphiploids
Benavente
E., Alix K. ,
Dusautoir J.-C., Orellana J.
and David J.L. 2001.
Early evolution of the chromosomal structure of Triticum turgidum -
Aegilops ovata amphiploids carrying and lacking the Ph1 gene. Theoretical and Applied Genetics
103: 1123-1128.
-
1995-1998:
PhD student, Agetrop,
CIRAD, Montpellier - J.-C. Glaszmann, A. D'Hont
The repetitive sequences of
sugarcane: genome-specificities and phylogeny of the
Saccharum
complex
1. Alix K., Paulet F., Glaszmann
J.-C. and D’Hont A. 1999. Inter-Alu-like
species-specific sequences in the Saccharum
complex. Theoretical and Applied
Genetics 99:
962-968.
2. Alix K., Baurens F.-C., Paulet F., Glaszmann J.-C. and D’Hont A. 1998. Isolation and
characterization of a satellite DNA family in the Saccharum complex.
Genome 41:
854-864.
3. D’Hont A., Ison D., Alix K., Roux C. and Glaszmann J.-C. 1998.
Determination of basic chromosome numbers in the genus Saccharum by physical mapping of
ribosomal RNA genes. Genome 41:
221-225.
Selected Publications
- Vitte C., Fustier M.-A., Alix K. and Tenaillon M. 2014. The bright side of transposons in crop evolution. Briefings in Functional Genomics 13: 276-295.

- Sarilar V., Martinez Palacios P., Rousselet A., Ridel C., Falque M., Eber F., Chèvre A.-M., Joets J., Brabant P., Alix K.
2013. Allopolyploidy has a moderate impact on restructuring at three
contrasting transposable element insertion sites in resynthesized Brassica napus allotetraploids. New Phytologist 198: 593-604.

- Jenczewski E., Chèvre A.-M. and Alix K.
2013. Chromosomal and gene expression changes in Brassica
allopolyploids. In: Polyploid and Hybrid Genomics. Z.J. Chen and J.A.
Birchler (Eds). John Wiley & Sons, Inc., Oxford, UK.

- Sarilar V., Marmagne A., Brabant P., Joets J., Alix K. 2011. BraSto, a Stowaway MITE from Brassica: recently active copies preferentially accumulate in the gene space. Plant Molecular Biology 77: 59-75.

- Marmagne A., Brabant P., Thiellement H. and Alix K. 2010. Analysis of gene expression in resynthesized Brassica napus allotetraploids: transcriptional changes do not explain differential protein regulation. New Phytologist 186: 216-227.

- Parisod C., Alix K.,
Just J., Petit M., Sarilar V., Mhiri C., Aïnouche M., Chalhoub B.
and Grandbastien M.-A. 2010. Impact of transposable elements on the
organization and funtion of allopolyploid genomes. New Phytologist 186: 37-45.

- Zerjal T., Joets J., Alix K.,
Grandbastien M.-A. and Tenaillon M. 2009. Contrasting evolutionary
patterns and target specificities among three Tourist-like MITE
families in the maize genome. Plant Molecular Biology 71: 99-114.

- Alix K.
, Joets J., Ryder C.D., Moore J., Barker G.C., Bailey J.P.,
King G.J. and Heslop-Harrison J.S. 2008. The CACTA transposon
Bot1
played a major role in
Brassica
genome divergence and gene proliferation.
The Plant Journal 56: 1030-1044.

- Pouilly N., Delourme R.,
Alix K.
and Jenczewski E. 2008. Repetitive sequence-derived markers tag
centromeres and telomeres and provide insights into chromosome
evolution in
Brassica napus
.
Chromosome Research
16: 683-700.

- Albertin W., Alix K.,
Balliau T., Brabant P., Davanture M., Malosse C., Valot B. and
Thiellement H. 2007. Differential regulation of gene products in newly
synthesized Brassica napus allotetraploids is not related to protein function nor subcellular localization. BMC Genomics 8: 56.

- Alix
K., Ryder C.D., Moore J.,
King G.J. and Heslop-Harrison J.S.
2005. The genomic organization
of retrotransposons in Brassica oleracea.
Plant Molecular
Biology 59:
839-851.

- Alix
K.
and Heslop-Harrison J.S. 2004. The diversity of retroelements in diploid
and allotetraploid Brassica species. Plant Molecular
Biology 54:
895-909.

- Jenczewski
E. and Alix K.
2004.
From diploids to allopolyploids: the emergence of efficient pairing control
genes in plants. Critical Reviews in Plant Sciences
23: 21-45.

Teaching at AgroParisTech
UFR Génétique évolutive et amélioration des plantes / Evolutionary genetics and plant breeding
First year
- Génétique des populations / Population genetics
- Enjeux et défis « Biodiversité sauvage et domestique : un état des lieux » / Stakes and challenges « the wild biodiversity and the cultivated biodiversity: the current situation »
- module intégratif « Gènes et Nature : les biotechnologies végétales au centre des questions de société»/«Genes and Nature: when green technologies meet Society »
Second year
- Socle commun du domaine 1 Productions, filières, territoires pour le développement durable, « Génétique et amélioration des plantes »/ « Plant genetics and breeding »
- Modules d’approfondissement : (i) « Ce que nous apprennent les génomes » / « What we can learn from genome exploration », (ii) « Adaptation des plantes à l'environnement » / « Adaptation of plants to environment », (iii) « Les défis de l'amélioration des plantes » /« The new challenges of plant breeding », (iv) « Biotechnologies, alimentation et santé »/« Biotechnologies, food and health »
« Initiation à la Recherche en Biologie » /« Discovering Research in Biology ». I had the pleasure to supervise the project of two students
of mine from AgroParisTech (from April to June 2013). They had to write
posts for AoB Blog, and produced several articles dealing with research
topics in plant biology. I invite you to visit AoB blog, and to have a
look at Aurélien Azam and Antoine Legal's posts. Enjoy reading
AoB blog!
Third year
- DA
Production et Innovation dans les Systèmes Techniques
Végétaux (PISTv) / Production and Innovation
in Plant Systems:
- Module « Amélioration des plantes :
méthodes, objectifs et acteurs de la filière semences
» / « Plant breeding: methods, aims and breeders »
- Module « Génomique, biotechnologies
végétales et biologie des semences » /« Genomics, plant biotechnologies and seed biology »
- Option de spécialisation « Amélioration des plantes
» /« Plant breeding »
- DA Biotechs:
- Module « Génomique et bioanalyse » / « Genomics and bioinformatics »
Teaching for Master students, Université Paris-Saclay 
Master 1, Biologie Intégrative et Physiologie (BIP)
- Socle commun : « Evolution et organisation des génomes
» / « Genome evolution and organisation
»
- Parcours végétal : (i) « Adaptation des plantes à l'environnement » / « Adaptation of plants to environment », (ii) « Génomique des espèces cultivées et bioinformatique
» / « Crop genomics and bioinformatics
», (iii) « Génétique appliquée à l'agronomie et la sélection variétale
» / « Genetics for agronomy and plant breeding
»
Master 2, Sciences du Végétal (ScV)
UE « Génomique et amélioration des plantes
» /« Genomics and plant breeding
»
Conferences and Others
I would be pleased to welcome you at AgroParisTech, Paris, for the next INRA workshop series DynaGeV, Dynamique des Génomes Végétaux / Plant Genome Dynamics, 7-8 July 2016. More information soon...
|
|



 ©Karine ALIX
©Karine ALIX


